- Scikit Image – Introduction
- Scikit Image - Image Processing
- Scikit Image - Numpy Images
- Scikit Image - Image datatypes
- Scikit Image - Using Plugins
- Scikit Image - Image Handlings
- Scikit Image - Reading Images
- Scikit Image - Writing Images
- Scikit Image - Displaying Images
- Scikit Image - Image Collections
- Scikit Image - Image Stack
- Scikit Image - Multi Image
- Scikit Image - Data Visualization
- Scikit Image - Using Matplotlib
- Scikit Image - Using Ploty
- Scikit Image - Using Mayavi
- Scikit Image - Using Napari
- Scikit Image - Color Manipulation
- Scikit Image - Alpha Channel
- Scikit Image - Conversion b/w Color & Gray Values
- Scikit Image - Conversion b/w RGB & HSV
- Scikit Image - Conversion to CIE-LAB Color Space
- Scikit Image - Conversion from CIE-LAB Color Space
- Scikit Image - Conversion to luv Color Space
- Scikit Image - Conversion from luv Color Space
- Scikit Image - Image Inversion
- Scikit Image - Painting Images with Labels
- Scikit Image - Contrast & Exposure
- Scikit Image - Contrast
- Scikit Image - Contrast enhancement
- Scikit Image - Exposure
- Scikit Image - Histogram Matching
- Scikit Image - Histogram Equalization
- Scikit Image - Local Histogram Equalization
- Scikit Image - Tinting gray-scale images
- Scikit Image - Image Transformation
- Scikit Image - Scaling an image
- Scikit Image - Rotating an Image
- Scikit Image - Warping an Image
- Scikit Image - Affine Transform
- Scikit Image - Piecewise Affine Transform
- Scikit Image - ProjectiveTransform
- Scikit Image - EuclideanTransform
- Scikit Image - Radon Transform
- Scikit Image - Line Hough Transform
- Scikit Image - Probabilistic Hough Transform
- Scikit Image - Circular Hough Transforms
- Scikit Image - Elliptical Hough Transforms
- Scikit Image - Polynomial Transform
- Scikit Image - Image Pyramids
- Scikit Image - Pyramid Gaussian Transform
- Scikit Image - Pyramid Laplacian Transform
- Scikit Image - Swirl Transform
- Scikit Image - Morphological Operations
- Scikit Image - Erosion
- Scikit Image - Dilation
- Scikit Image - Black & White Tophat Morphologies
- Scikit Image - Convex Hull
- Scikit Image - Generating footprints
- Scikit Image - Isotopic Dilation & Erosion
- Scikit Image - Isotopic Closing & Opening of an Image
- Scikit Image - Skelitonizing an Image
- Scikit Image - Morphological Thinning
- Scikit Image - Masking an image
- Scikit Image - Area Closing & Opening of an Image
- Scikit Image - Diameter Closing & Opening of an Image
- Scikit Image - Morphological reconstruction of an Image
- Scikit Image - Finding local Maxima
- Scikit Image - Finding local Minima
- Scikit Image - Removing Small Holes from an Image
- Scikit Image - Removing Small Objects from an Image
- Scikit Image - Filters
- Scikit Image - Image Filters
- Scikit Image - Median Filter
- Scikit Image - Mean Filters
- Scikit Image - Morphological gray-level Filters
- Scikit Image - Gabor Filter
- Scikit Image - Gaussian Filter
- Scikit Image - Butterworth Filter
- Scikit Image - Frangi Filter
- Scikit Image - Hessian Filter
- Scikit Image - Meijering Neuriteness Filter
- Scikit Image - Sato Filter
- Scikit Image - Sobel Filter
- Scikit Image - Farid Filter
- Scikit Image - Scharr Filter
- Scikit Image - Unsharp Mask Filter
- Scikit Image - Roberts Cross Operator
- Scikit Image - Lapalace Operator
- Scikit Image - Window Functions With Images
- Scikit Image - Thresholding
- Scikit Image - Applying Threshold
- Scikit Image - Otsu Thresholding
- Scikit Image - Local thresholding
- Scikit Image - Hysteresis Thresholding
- Scikit Image - Li thresholding
- Scikit Image - Multi-Otsu Thresholding
- Scikit Image - Niblack and Sauvola Thresholding
- Scikit Image - Restoring Images
- Scikit Image - Rolling-ball Algorithm
- Scikit Image - Denoising an Image
- Scikit Image - Wavelet Denoising
- Scikit Image - Non-local means denoising for preserving textures
- Scikit Image - Calibrating Denoisers Using J-Invariance
- Scikit Image - Total Variation Denoising
- Scikit Image - Shift-invariant wavelet denoising
- Scikit Image - Image Deconvolution
- Scikit Image - Richardson-Lucy Deconvolution
- Scikit Image - Recover the original from a wrapped phase image
- Scikit Image - Image Inpainting
- Scikit Image - Registering Images
- Scikit Image - Image Registration
- Scikit Image - Masked Normalized Cross-Correlation
- Scikit Image - Registration using optical flow
- Scikit Image - Assemble images with simple image stitching
- Scikit Image - Registration using Polar and Log-Polar
- Scikit Image - Feature Detection
- Scikit Image - Dense DAISY Feature Description
- Scikit Image - Histogram of Oriented Gradients
- Scikit Image - Template Matching
- Scikit Image - CENSURE Feature Detector
- Scikit Image - BRIEF Binary Descriptor
- Scikit Image - SIFT Feature Detector and Descriptor Extractor
- Scikit Image - GLCM Texture Features
- Scikit Image - Shape Index
- Scikit Image - Sliding Window Histogram
- Scikit Image - Finding Contour
- Scikit Image - Texture Classification Using Local Binary Pattern
- Scikit Image - Texture Classification Using Multi-Block Local Binary Pattern
- Scikit Image - Active Contour Model
- Scikit Image - Canny Edge Detection
- Scikit Image - Marching Cubes
- Scikit Image - Foerstner Corner Detection
- Scikit Image - Harris Corner Detection
- Scikit Image - Extracting FAST Corners
- Scikit Image - Shi-Tomasi Corner Detection
- Scikit Image - Haar Like Feature Detection
- Scikit Image - Haar Feature detection of coordinates
- Scikit Image - Hessian matrix
- Scikit Image - ORB feature Detection
- Scikit Image - Additional Concepts
- Scikit Image - Render text onto an image
- Scikit Image - Face detection using a cascade classifier
- Scikit Image - Face classification using Haar-like feature descriptor
- Scikit Image - Visual image comparison
- Scikit Image - Exploring Region Properties With Pandas
Recover the Original from a Wrapped Phase Image
The process of recovering the original from a wrapped phase image typically involves a technique called "phase unwrapping. The phase unwrapping method aims to retrieve the underlying, continuous, or unwrapped phase information from an image or signal that has undergone a wrapping process.
In some situations, certain signals or images are restricted to values within a limited range (modulo 2*pi). In these cases, phase unwrapping is used to recover the underlying, unwrapped signal. In this tutorial, we'll see about the scikit-image (skimage) provided phase unwrapping algorithm on one-dimensional, two-dimensional, and three-dimensional images.
Using the skimage.restoration.unwrap_phase() function
The restoration.unwrap_phase() function is used to recover the original, unwrapped image from a wrapped phase image. It recovers an image from a wrapped image having values that lie in the interval [-pi, pi), and recovers the original, unwrapped image.
Syntax
Following is the syntax of this function −
skimage.restoration.unwrap_phase(image, wrap_around=False, rng=None)
Parameters
Here are the details about the function parameters −
image: This is a 1D, 2D, or 3D ndarray of floats, optionally a masked array. It represents the wrapped phase image and values in this image should be in the range [-pi, pi). If a masked array is provided, the masked entries will not be changed, and their values will not be used to guide the unwrapping of neighboring, unmasked values. However, masked 1D arrays are not allowed and will raise a ValueError.
wrap_around (optional): This parameter is a boolean or a sequence of booleans. When an element of the sequence is True, the unwrapping process will regard the edges along the corresponding axis of the image to be connected and use this connectivity to guide the phase unwrapping process. If only a single boolean is given, it will apply to all axes. This wrap-around is not supported for 1D arrays.
rng (optional): This parameter specifies the pseudo-random number generator used for initialization during the unwrapping process. By default, a PCG64 generator is used (see numpy.random.default_rng()). If rng is an integer, it is used to seed the generator. Setting a specific seed can be useful for achieving deterministic behavior in the unwrapping process.
The function returns an unwrapped image (image_unwrapped: array_like, double) of the same shape as the input. If the input image is a masked array, the mask information will be preserved in the output.
Additionally, the function can raise a ValueError in the following cases −
If called with a masked 1D array.
If called with a 1D array and wrap_around=True.
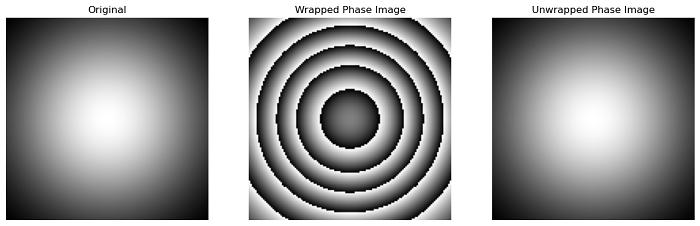
Example
The following example demonstrates how to use the unwrap_phase() function to unwrap a wrapped phase image.
import numpy as np
import matplotlib.pyplot as plt
from skimage.restoration import unwrap_phase
# Create the coordinate grids
c0, c1 = np.ogrid[-1:1:128j, -1:1:128j]
# Create the input wrapped phase image
image = 12 * np.pi * np.exp(-(c0**2 + c1**2))
image_wrapped = np.angle(np.exp(1j * image))
# Unwrap the phase image
image_unwrapped = unwrap_phase(image_wrapped)
# Create subplots for the display the results
fig, axes = plt.subplots(1, 3, figsize=(15, 5))
ax = axes.ravel()
plt.gray()
# Display the original image
ax[0].imshow(image)
ax[0].set_title('Original')
ax[0].axis('off')
# Display the Wrapped Phase Image
ax[1].imshow(image_wrapped)
ax[1].set_title('Wrapped Phase Image')
ax[1].axis('off')
# Unwrapped Phase Image
ax[2].imshow(image_unwrapped)
ax[2].set_title('Unwrapped Phase Image')
ax[2].axis('off')
plt.show()
Output
Example
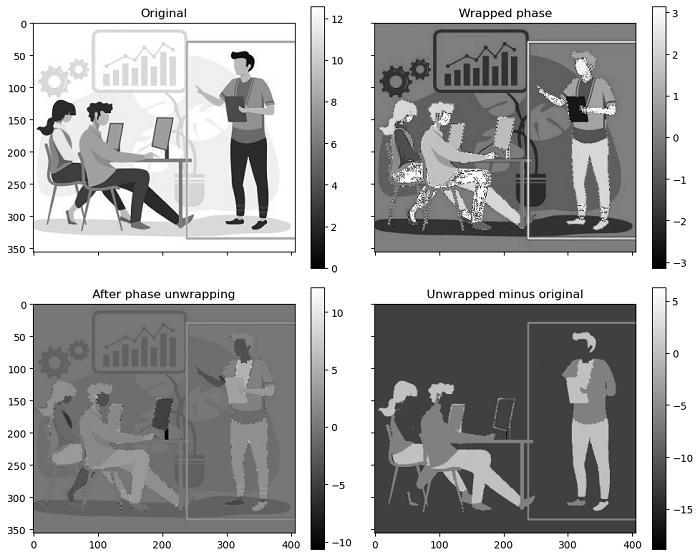
The following example demonstrates the process of phase unwrapping using the unwrap_phase() function of a two dimensional wrapped phase image.
import numpy as np
import matplotlib.pyplot as plt
from skimage import color, io, exposure
from skimage.restoration import unwrap_phase
# Load an image as a floating-point grayscale
original_image = color.rgb2gray(io.imread('Images/image5.jpg'))
# Scale the image to the range [0, 4*pi]
scaled_image = exposure.rescale_intensity(original_image, out_range=(0, 4 * np.pi))
# Create a phase-wrapped image in the interval [-pi, pi)
wrapped_phase_image = np.angle(np.exp(1j * scaled_image))
# Perform phase unwrapping
unwrapped_phase_image = unwrap_phase(wrapped_phase_image)
# Create subplots for visualization
fig, axes = plt.subplots(2, 2, sharex=True, sharey=True)
ax1, ax2, ax3, ax4 = axes.ravel()
# Display the original image
ax1.imshow(scaled_image, cmap='gray', vmin=0, vmax=4 * np.pi)
ax1.set_title('Original')
# Display the wrapped phase image
ax2.imshow(wrapped_phase_image, cmap='gray', vmin=-np.pi, vmax=np.pi)
ax2.set_title('Wrapped phase')
# Display the unwrapped phase image
ax3.imshow(unwrapped_phase_image, cmap='gray')
ax3.set_title('After phase unwrapping')
# Display the difference between unwrapped and original
ax4.imshow(unwrapped_phase_image - scaled_image, cmap='gray')
ax4.set_title('Unwrapped minus original')
# Add colorbars for reference
for ax in axes.ravel():
fig.colorbar(ax.get_images()[0], ax=ax)
plt.tight_layout()
plt.show()
Output
Unwrapping with Masked Arrays
The unwrapping procedure (unwrap_phase() function) can accept masked arrays as input. When a masked array is provided, the values in the masked entries are not changed during the unwrapping process. Additionally, the values of masked entries are not used to guide the unwrapping of neighboring unmasked values.
Example
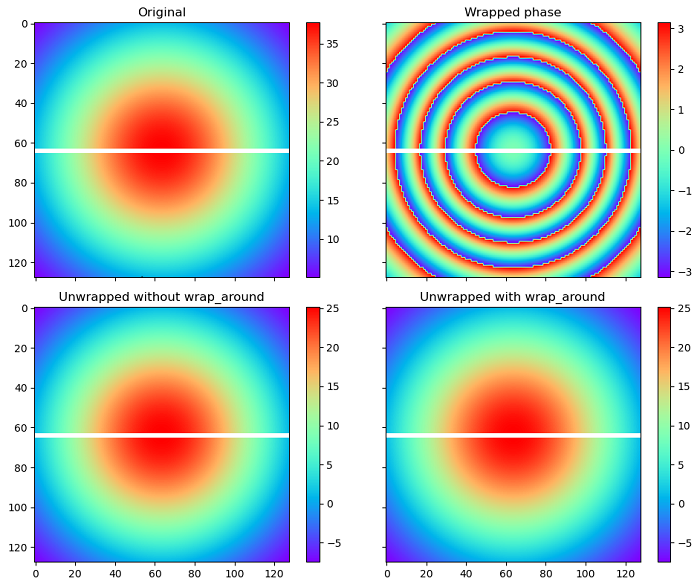
Here is an example that demonstrates the unwrapping of the wrapped phase image with a mask using the unwrap_phase() function with and without wrap-around enabled.
import numpy as np
import matplotlib.pyplot as plt
from skimage.restoration import unwrap_phase
# Create the coordinate grids
c0, c1 = np.ogrid[-1:1:128j, -1:1:128j]
# Create the input image
image = 12 * np.pi * np.exp(-(c0**2 + c1**2))
# Mask the image to split it in two horizontally
mask = np.zeros_like(image, dtype=bool)
mask[image.shape[0] // 2, :] = True
# Compute the wrapped phase image
image_wrapped = np.ma.array(np.angle(np.exp(1j * image)), mask=mask)
# Unwrap image without wrap around
image_unwrapped_no_wrap_around = unwrap_phase(image_wrapped, wrap_around=(False, False))
# Unwrap with wrap around enabled for the 0th dimension
image_unwrapped_wrap_around = unwrap_phase(image_wrapped, wrap_around=(True, False))
# Create subplots for visualization
fig, ax = plt.subplots(2, 2, figsize=(10, 8),sharex=True, sharey=True)
ax1, ax2, ax3, ax4 = ax.ravel()
# Plot the original image with the mask
fig.colorbar(ax1.imshow(np.ma.array(image, mask=mask), cmap='rainbow'), ax=ax1)
ax1.set_title('Original')
# Plot the wrapped phase image
fig.colorbar(ax2.imshow(image_wrapped, cmap='rainbow', vmin=-np.pi, vmax=np.pi), ax=ax2)
ax2.set_title('Wrapped phase')
# Plot the unwrapped image without wrap_around
fig.colorbar(ax3.imshow(image_unwrapped_no_wrap_around, cmap='rainbow'), ax=ax3)
ax3.set_title('Unwrapped without wrap_around')
# Plot the unwrapped image with wrap_around
fig.colorbar(ax4.imshow(image_unwrapped_wrap_around, cmap='rainbow'), ax=ax4)
ax4.set_title('Unwrapped with wrap_around')
plt.tight_layout()
plt.show()
Output