- Scikit Image – Introduction
- Scikit Image - Image Processing
- Scikit Image - Numpy Images
- Scikit Image - Image datatypes
- Scikit Image - Using Plugins
- Scikit Image - Image Handlings
- Scikit Image - Reading Images
- Scikit Image - Writing Images
- Scikit Image - Displaying Images
- Scikit Image - Image Collections
- Scikit Image - Image Stack
- Scikit Image - Multi Image
- Scikit Image - Data Visualization
- Scikit Image - Using Matplotlib
- Scikit Image - Using Ploty
- Scikit Image - Using Mayavi
- Scikit Image - Using Napari
- Scikit Image - Color Manipulation
- Scikit Image - Alpha Channel
- Scikit Image - Conversion b/w Color & Gray Values
- Scikit Image - Conversion b/w RGB & HSV
- Scikit Image - Conversion to CIE-LAB Color Space
- Scikit Image - Conversion from CIE-LAB Color Space
- Scikit Image - Conversion to luv Color Space
- Scikit Image - Conversion from luv Color Space
- Scikit Image - Image Inversion
- Scikit Image - Painting Images with Labels
- Scikit Image - Contrast & Exposure
- Scikit Image - Contrast
- Scikit Image - Contrast enhancement
- Scikit Image - Exposure
- Scikit Image - Histogram Matching
- Scikit Image - Histogram Equalization
- Scikit Image - Local Histogram Equalization
- Scikit Image - Tinting gray-scale images
- Scikit Image - Image Transformation
- Scikit Image - Scaling an image
- Scikit Image - Rotating an Image
- Scikit Image - Warping an Image
- Scikit Image - Affine Transform
- Scikit Image - Piecewise Affine Transform
- Scikit Image - ProjectiveTransform
- Scikit Image - EuclideanTransform
- Scikit Image - Radon Transform
- Scikit Image - Line Hough Transform
- Scikit Image - Probabilistic Hough Transform
- Scikit Image - Circular Hough Transforms
- Scikit Image - Elliptical Hough Transforms
- Scikit Image - Polynomial Transform
- Scikit Image - Image Pyramids
- Scikit Image - Pyramid Gaussian Transform
- Scikit Image - Pyramid Laplacian Transform
- Scikit Image - Swirl Transform
- Scikit Image - Morphological Operations
- Scikit Image - Erosion
- Scikit Image - Dilation
- Scikit Image - Black & White Tophat Morphologies
- Scikit Image - Convex Hull
- Scikit Image - Generating footprints
- Scikit Image - Isotopic Dilation & Erosion
- Scikit Image - Isotopic Closing & Opening of an Image
- Scikit Image - Skelitonizing an Image
- Scikit Image - Morphological Thinning
- Scikit Image - Masking an image
- Scikit Image - Area Closing & Opening of an Image
- Scikit Image - Diameter Closing & Opening of an Image
- Scikit Image - Morphological reconstruction of an Image
- Scikit Image - Finding local Maxima
- Scikit Image - Finding local Minima
- Scikit Image - Removing Small Holes from an Image
- Scikit Image - Removing Small Objects from an Image
- Scikit Image - Filters
- Scikit Image - Image Filters
- Scikit Image - Median Filter
- Scikit Image - Mean Filters
- Scikit Image - Morphological gray-level Filters
- Scikit Image - Gabor Filter
- Scikit Image - Gaussian Filter
- Scikit Image - Butterworth Filter
- Scikit Image - Frangi Filter
- Scikit Image - Hessian Filter
- Scikit Image - Meijering Neuriteness Filter
- Scikit Image - Sato Filter
- Scikit Image - Sobel Filter
- Scikit Image - Farid Filter
- Scikit Image - Scharr Filter
- Scikit Image - Unsharp Mask Filter
- Scikit Image - Roberts Cross Operator
- Scikit Image - Lapalace Operator
- Scikit Image - Window Functions With Images
- Scikit Image - Thresholding
- Scikit Image - Applying Threshold
- Scikit Image - Otsu Thresholding
- Scikit Image - Local thresholding
- Scikit Image - Hysteresis Thresholding
- Scikit Image - Li thresholding
- Scikit Image - Multi-Otsu Thresholding
- Scikit Image - Niblack and Sauvola Thresholding
- Scikit Image - Restoring Images
- Scikit Image - Rolling-ball Algorithm
- Scikit Image - Denoising an Image
- Scikit Image - Wavelet Denoising
- Scikit Image - Non-local means denoising for preserving textures
- Scikit Image - Calibrating Denoisers Using J-Invariance
- Scikit Image - Total Variation Denoising
- Scikit Image - Shift-invariant wavelet denoising
- Scikit Image - Image Deconvolution
- Scikit Image - Richardson-Lucy Deconvolution
- Scikit Image - Recover the original from a wrapped phase image
- Scikit Image - Image Inpainting
- Scikit Image - Registering Images
- Scikit Image - Image Registration
- Scikit Image - Masked Normalized Cross-Correlation
- Scikit Image - Registration using optical flow
- Scikit Image - Assemble images with simple image stitching
- Scikit Image - Registration using Polar and Log-Polar
- Scikit Image - Feature Detection
- Scikit Image - Dense DAISY Feature Description
- Scikit Image - Histogram of Oriented Gradients
- Scikit Image - Template Matching
- Scikit Image - CENSURE Feature Detector
- Scikit Image - BRIEF Binary Descriptor
- Scikit Image - SIFT Feature Detector and Descriptor Extractor
- Scikit Image - GLCM Texture Features
- Scikit Image - Shape Index
- Scikit Image - Sliding Window Histogram
- Scikit Image - Finding Contour
- Scikit Image - Texture Classification Using Local Binary Pattern
- Scikit Image - Texture Classification Using Multi-Block Local Binary Pattern
- Scikit Image - Active Contour Model
- Scikit Image - Canny Edge Detection
- Scikit Image - Marching Cubes
- Scikit Image - Foerstner Corner Detection
- Scikit Image - Harris Corner Detection
- Scikit Image - Extracting FAST Corners
- Scikit Image - Shi-Tomasi Corner Detection
- Scikit Image - Haar Like Feature Detection
- Scikit Image - Haar Feature detection of coordinates
- Scikit Image - Hessian matrix
- Scikit Image - ORB feature Detection
- Scikit Image - Additional Concepts
- Scikit Image - Render text onto an image
- Scikit Image - Face detection using a cascade classifier
- Scikit Image - Face classification using Haar-like feature descriptor
- Scikit Image - Visual image comparison
- Scikit Image - Exploring Region Properties With Pandas
Scikit Image - Marching Cubes
Marching cubes is a 3D algorithm used to create a surface mesh from a 3D volume. This can be conceptualized as a 3D generalization of isolines on topographical or weather maps. It works by iterating through the volume, identifying regions that meet a certain level of interest, and then creating triangles to build a 3D mesh. This algorithm requires two main inputs: the 3D data volume and an isosurface value. For instance, in CT imaging Hounsfield units of +700 to +3000 represent bone density within the CT data. So, one potential input would be a reconstructed CT set of data and the value +700, to extract a mesh for regions of bone or bone-like density. The result is a mesh consisting of vertices and triangular faces.
The scikit image library provides the marching_cubes() function within its measure module to work with 3D volumetric data.
Using the skimage.measure.marching_cubes() function
The marching_cubes() function is an implementation of the Marching Cubes algorithm designed to identify surfaces within 3D volumetric data. Unlike the approach by Lorensen et al., this implementation follows the algorithm by Lewiner et al. This choice brings several advantages: it's faster, resolves ambiguities, and ensures topologically correct results. Therefore, when working with 3D data, using marching_cubes() with the Lewiner et al. method is generally the better choice.
Syntax
Following is the syntax of this function −
skimage.measure.marching_cubes(volume, level=None, *, spacing=(1.0, 1.0, 1.0), gradient_direction='descent', step_size=1, allow_degenerate=True, method='lewiner', mask=None)
Parameters
The function accepts the following parameters −
volume (M, N, P) array: Input 3D data volume from which isosurfaces will be extracted. It Will internally be converted to float32 if necessary.
level (float, optional): Contour value to search for isosurfaces in the volume. Default is the average of the minimum and maximum values in the volume.
spacing (length-3 tuple of floats, optional): Voxel spacing in spatial dimensions corresponding to numpy array indexing dimensions (M, N, P) as in the volume.
gradient_direction (string, optional): Specifies whether the mesh is generated from an isosurface with gradient descent or ascent.
step_size (int, optional): Step size in voxels. The default value is 1. Larger steps yield faster but coarser results. The result will always be topologically correct.
allow_degenerate (bool, optional): Determines whether degenerate (i.e. zero-area) triangles are allowed in the output mesh. The default is True. If False, degenerate triangles are removed, making the algorithm slower.
method ({'lewiner', 'lorensen'}, optional): Specifies the algorithm to use, either Lewiner et al. or the Lorensen et al. method.
mask (M, N, P) array, optional: Boolean array to restrict the computation to certain regions of the volume. The marching cube algorithm will be computed only on True elements, saving computational time and allowing for finite surfaces.
Function returns −
verts (V, 3) array: Spatial coordinates for V unique mesh vertices. Coordinate order matches the input volume (M, N, P).
faces (F, 3) array: Defines triangular faces via referencing vertex indices from verts. Each face has exactly three indices.
normals (V, 3) array: The normal direction at each vertex, as calculated from the data.
values (V, ) array: Gives a measure for the maximum value of the data in the local region near each vertex, useful for applying a colormap to the mesh in visualization tools.
Example
The following example demonstrates how to use the Marching Cubes algorithm to generate and visualize a 3D mesh from a 3D dataset.
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d.art3d import Poly3DCollection
from skimage import measure
from skimage.draw import ellipsoid
# Generate a level set of two identical ellipsoids in 3D
ellipsoid_base = ellipsoid(6, 10, 16, levelset=True)
# Create a larger dataset by stacking two identical ellipsoids along the z-axis
ellipsoid_double = np.concatenate((ellipsoid_base[:-1, ...], ellipsoid_base[2:, ...]), axis=0)
# Extract the surface mesh of the ellipsoids using marching cubes
vertices, faces, normals, values = measure.marching_cubes(ellipsoid_double, 0)
# Create a 3D plot for visualization
fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(111, projection='3d')
# Create a collection of triangles from vertices and faces
mesh = Poly3DCollection(vertices[faces])
mesh.set_edgecolor('k')
ax.add_collection3d(mesh)
# Set labels and axis limits
ax.set_xlabel("X-axis: a = 6 per ellipsoid")
ax.set_ylabel("Y-axis: b = 10")
ax.set_zlabel("Z-axis: c = 16")
ax.set_xlim(0, 24)
ax.set_ylim(0, 20)
ax.set_zlim(0, 32)
plt.tight_layout()
plt.show()
Output
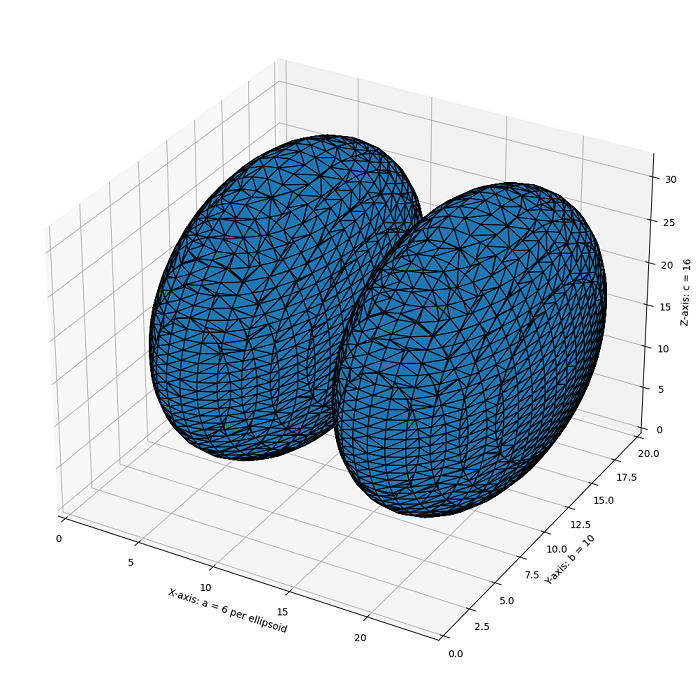
Example
Here is an example that demonstrates how to visualize the output of the Marching Cubes algorithm using the Mayavi package.
from mayavi import mlab from skimage.measure import marching_cubes import matplotlib.pyplot as plt from skimage import measure from skimage.draw import ellipsoid # Generate an identical ellipsoids in 3D ellipsoid_base = ellipsoid(6, 10, 16, levelset=True) # Extract the surface mesh of the ellipsoids using marching cubes verts, faces, _, _ = measure.marching_cubes(ellipsoid_base, 0.0) # Extract coordinates from vertices x_coords = [vert[0] for vert in verts] y_coords = [vert[1] for vert in verts] z_coords = [vert[2] for vert in verts] # Create a 3D triangular mesh using Mayavi mlab.triangular_mesh(x_coords, y_coords, z_coords, faces) # Display the 3D mesh mlab.show()
Output