
- Scikit Image – Introduction
- Scikit Image - Image Processing
- Scikit Image - Numpy Images
- Scikit Image - Image datatypes
- Scikit Image - Using Plugins
- Scikit Image - Image Handlings
- Scikit Image - Reading Images
- Scikit Image - Writing Images
- Scikit Image - Displaying Images
- Scikit Image - Image Collections
- Scikit Image - Image Stack
- Scikit Image - Multi Image
- Scikit Image - Data Visualization
- Scikit Image - Using Matplotlib
- Scikit Image - Using Ploty
- Scikit Image - Using Mayavi
- Scikit Image - Using Napari
- Scikit Image - Color Manipulation
- Scikit Image - Alpha Channel
- Scikit Image - Conversion b/w Color & Gray Values
- Scikit Image - Conversion b/w RGB & HSV
- Scikit Image - Conversion to CIE-LAB Color Space
- Scikit Image - Conversion from CIE-LAB Color Space
- Scikit Image - Conversion to luv Color Space
- Scikit Image - Conversion from luv Color Space
- Scikit Image - Image Inversion
- Scikit Image - Painting Images with Labels
- Scikit Image - Contrast & Exposure
- Scikit Image - Contrast
- Scikit Image - Contrast enhancement
- Scikit Image - Exposure
- Scikit Image - Histogram Matching
- Scikit Image - Histogram Equalization
- Scikit Image - Local Histogram Equalization
- Scikit Image - Tinting gray-scale images
- Scikit Image - Image Transformation
- Scikit Image - Scaling an image
- Scikit Image - Rotating an Image
- Scikit Image - Warping an Image
- Scikit Image - Affine Transform
- Scikit Image - Piecewise Affine Transform
- Scikit Image - ProjectiveTransform
- Scikit Image - EuclideanTransform
- Scikit Image - Radon Transform
- Scikit Image - Line Hough Transform
- Scikit Image - Probabilistic Hough Transform
- Scikit Image - Circular Hough Transforms
- Scikit Image - Elliptical Hough Transforms
- Scikit Image - Polynomial Transform
- Scikit Image - Image Pyramids
- Scikit Image - Pyramid Gaussian Transform
- Scikit Image - Pyramid Laplacian Transform
- Scikit Image - Swirl Transform
- Scikit Image - Morphological Operations
- Scikit Image - Erosion
- Scikit Image - Dilation
- Scikit Image - Black & White Tophat Morphologies
- Scikit Image - Convex Hull
- Scikit Image - Generating footprints
- Scikit Image - Isotopic Dilation & Erosion
- Scikit Image - Isotopic Closing & Opening of an Image
- Scikit Image - Skelitonizing an Image
- Scikit Image - Morphological Thinning
- Scikit Image - Masking an image
- Scikit Image - Area Closing & Opening of an Image
- Scikit Image - Diameter Closing & Opening of an Image
- Scikit Image - Morphological reconstruction of an Image
- Scikit Image - Finding local Maxima
- Scikit Image - Finding local Minima
- Scikit Image - Removing Small Holes from an Image
- Scikit Image - Removing Small Objects from an Image
- Scikit Image - Filters
- Scikit Image - Image Filters
- Scikit Image - Median Filter
- Scikit Image - Mean Filters
- Scikit Image - Morphological gray-level Filters
- Scikit Image - Gabor Filter
- Scikit Image - Gaussian Filter
- Scikit Image - Butterworth Filter
- Scikit Image - Frangi Filter
- Scikit Image - Hessian Filter
- Scikit Image - Meijering Neuriteness Filter
- Scikit Image - Sato Filter
- Scikit Image - Sobel Filter
- Scikit Image - Farid Filter
- Scikit Image - Scharr Filter
- Scikit Image - Unsharp Mask Filter
- Scikit Image - Roberts Cross Operator
- Scikit Image - Lapalace Operator
- Scikit Image - Window Functions With Images
- Scikit Image - Thresholding
- Scikit Image - Applying Threshold
- Scikit Image - Otsu Thresholding
- Scikit Image - Local thresholding
- Scikit Image - Hysteresis Thresholding
- Scikit Image - Li thresholding
- Scikit Image - Multi-Otsu Thresholding
- Scikit Image - Niblack and Sauvola Thresholding
- Scikit Image - Restoring Images
- Scikit Image - Rolling-ball Algorithm
- Scikit Image - Denoising an Image
- Scikit Image - Wavelet Denoising
- Scikit Image - Non-local means denoising for preserving textures
- Scikit Image - Calibrating Denoisers Using J-Invariance
- Scikit Image - Total Variation Denoising
- Scikit Image - Shift-invariant wavelet denoising
- Scikit Image - Image Deconvolution
- Scikit Image - Richardson-Lucy Deconvolution
- Scikit Image - Recover the original from a wrapped phase image
- Scikit Image - Image Inpainting
- Scikit Image - Registering Images
- Scikit Image - Image Registration
- Scikit Image - Masked Normalized Cross-Correlation
- Scikit Image - Registration using optical flow
- Scikit Image - Assemble images with simple image stitching
- Scikit Image - Registration using Polar and Log-Polar
- Scikit Image - Feature Detection
- Scikit Image - Dense DAISY Feature Description
- Scikit Image - Histogram of Oriented Gradients
- Scikit Image - Template Matching
- Scikit Image - CENSURE Feature Detector
- Scikit Image - BRIEF Binary Descriptor
- Scikit Image - SIFT Feature Detector and Descriptor Extractor
- Scikit Image - GLCM Texture Features
- Scikit Image - Shape Index
- Scikit Image - Sliding Window Histogram
- Scikit Image - Finding Contour
- Scikit Image - Texture Classification Using Local Binary Pattern
- Scikit Image - Texture Classification Using Multi-Block Local Binary Pattern
- Scikit Image - Active Contour Model
- Scikit Image - Canny Edge Detection
- Scikit Image - Marching Cubes
- Scikit Image - Foerstner Corner Detection
- Scikit Image - Harris Corner Detection
- Scikit Image - Extracting FAST Corners
- Scikit Image - Shi-Tomasi Corner Detection
- Scikit Image - Haar Like Feature Detection
- Scikit Image - Haar Feature detection of coordinates
- Scikit Image - Hessian matrix
- Scikit Image - ORB feature Detection
- Scikit Image - Additional Concepts
- Scikit Image - Render text onto an image
- Scikit Image - Face detection using a cascade classifier
- Scikit Image - Face classification using Haar-like feature descriptor
- Scikit Image - Visual image comparison
- Scikit Image - Exploring Region Properties With Pandas
Scikit Image - Image Inpainting
In general, inpainting is a technique used for preserving and restoring artworks by filling in damaged, deteriorated, or missing portions to create a seamless image. In the context of image processing, inpainting involves the automatic reconstruction of lost or deteriorated sections of images and videos using information from undamaged areas. This process finds extensive use in image restoration.
The scikit-Image library offers an inpainting algorithm that operates based on the 'biharmonic equation'. This algorithm is available within the restoration module and is designed to efficiently perform inpainting tasks.
Using the skimage.restoration.inpaint_biharmonic() function
The restoration.inpaint_biharmonic() function is specifically designed for inpainting masked points within an image using biharmonic equations. It enables the reconstruction of missing or damaged portions by considering the surrounding information.
Syntax
Following is the syntax of this function −
skimage.restoration.inpaint_biharmonic(image, mask, *, split_into_regions=False, channel_axis=None)
Parameters
Here are the details of the parameters of the function −
image (M[, N[, , P]][, C]) ndarray): The input image that needs inpainting.
mask (M[, N[, , P]][, C]) ndarray): An array specifying the pixels to be inpainted. It should have the same shape as one of the 'image' channels. Unknown pixels should be represented with the value 1, while known pixels should be represented with the value 0.
split_into_regions (boolean, optional): If set to True, inpainting is done on a region-by-region basis. This approach may be slower but can reduce memory requirements.
channel_axis (int or None, optional): If set to None, the image is assumed to be grayscale (single channel). Otherwise, this parameter indicates which axis of the array corresponds to channels. This parameter was added from Scikit-Image version 0.19.
The function returns the inpainted image (out:(M[, N[, , P]][, C]) ndarray) with masked pixels filled in. It has the same shape as the original image.
Example
This example demonstrates the use of the inpaint_biharmonic() function to inpaint a damaged or missing region in an image based on a given mask.
import numpy as np
from skimage.restoration import inpaint_biharmonic
import matplotlib.pyplot as plt
# Create an example image
img = np.array([[ 1, 6, 3, 0, 4],
[ 1, 6, 3, 0, 4],
[ 1, 6, 3, 0, 4],
[ 1, 6, 3, 0, 4],
[ 1, 6, 3, 0, 4]], dtype='int32')
# Create a mask with known and unknown pixels
mask = np.zeros_like(img)
mask[2, 2:] = 1
mask[1, 3:] = 1
mask[0, 4:] = 1
mask[3, 0] = 1
mask[0, 0] = 1
# Perform inpainting using biharmonic equations
out = inpaint_biharmonic(img, mask)
# Visualize the original and inpainted images
fig, axes = plt.subplots(1, 3, figsize=(10, 5))
# Original Image
axes[0].imshow(img, cmap='gray')
axes[0].set_title('Original Image')
axes[0].axis('off')
# Mask
axes[1].imshow(mask, cmap='gray')
axes[1].set_title('Mask')
axes[1].axis('off')
# Inpainted Image
axes[2].imshow(out, cmap='gray')
axes[2].set_title('Inpainted Image')
axes[2].axis('off')
plt.tight_layout()
plt.show()
Output
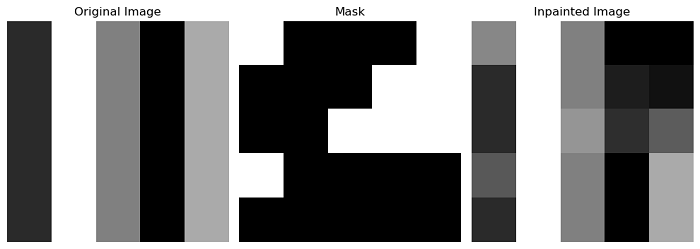
Example
This example demonstrates how the inpaint_biharmonic() function is used to restore the defected regions in the image.
import numpy as np
import matplotlib.pyplot as plt
from skimage import io
from skimage.morphology import disk, binary_dilation
from skimage.restoration import inpaint
# Load the input image
image = io.imread('Images/butterfly1.jpg')
# Create a mask with multiple defect regions
mask = np.zeros(image.shape[:-1], dtype=bool)
# Define and add six block defect regions
mask[20:60, 0:20] = 1
mask[160:180, 70:155] = 1
mask[30:60, 170:195] = 1
mask[-60:-30, 170:195] = 1
mask[-180:-160, 70:155] = 1
mask[-60:-20, 0:20] = 1
# Add a few long, narrow defects
mask[200:205, -200:] = 1
mask[150:255, 20:23] = 1
mask[365:368, 60:130] = 1
# Add randomly positioned small point-like defects
rstate = np.random.default_rng(0)
for radius in [0, 2, 4]:
# Larger defects are less common
# Make larger defects less common
thresh = 3 + 0.25 * radius
tmp_mask = rstate.standard_normal(image.shape[:-1]) > thresh
if radius > 0:
tmp_mask = binary_dilation(tmp_mask, disk(radius, dtype=bool))
mask[tmp_mask] = 1
# Apply the defect mask to the original image to create a defected image
image_defect = image * ~mask[..., np.newaxis]
# Use inpainting to restore the defected regions in the image
image_result = inpaint.inpaint_biharmonic(image_defect, mask, channel_axis=-1)
# Create subplots for visualization
fig, axes = plt.subplots(ncols=2, nrows=2, figsize=(10,8))
ax = axes.ravel()
ax[0].set_title('Original image')
ax[0].imshow(image)
ax[1].set_title('Mask')
ax[1].imshow(mask, cmap=plt.cm.gray)
ax[2].set_title('Defected image')
ax[2].imshow(image_defect)
ax[3].set_title('Inpainted image')
ax[3].imshow(image_result)
for a in ax:
a.axis('off')
fig.tight_layout()
plt.show()
Output
