- Scikit Image – Introduction
- Scikit Image - Image Processing
- Scikit Image - Numpy Images
- Scikit Image - Image datatypes
- Scikit Image - Using Plugins
- Scikit Image - Image Handlings
- Scikit Image - Reading Images
- Scikit Image - Writing Images
- Scikit Image - Displaying Images
- Scikit Image - Image Collections
- Scikit Image - Image Stack
- Scikit Image - Multi Image
- Scikit Image - Data Visualization
- Scikit Image - Using Matplotlib
- Scikit Image - Using Ploty
- Scikit Image - Using Mayavi
- Scikit Image - Using Napari
- Scikit Image - Color Manipulation
- Scikit Image - Alpha Channel
- Scikit Image - Conversion b/w Color & Gray Values
- Scikit Image - Conversion b/w RGB & HSV
- Scikit Image - Conversion to CIE-LAB Color Space
- Scikit Image - Conversion from CIE-LAB Color Space
- Scikit Image - Conversion to luv Color Space
- Scikit Image - Conversion from luv Color Space
- Scikit Image - Image Inversion
- Scikit Image - Painting Images with Labels
- Scikit Image - Contrast & Exposure
- Scikit Image - Contrast
- Scikit Image - Contrast enhancement
- Scikit Image - Exposure
- Scikit Image - Histogram Matching
- Scikit Image - Histogram Equalization
- Scikit Image - Local Histogram Equalization
- Scikit Image - Tinting gray-scale images
- Scikit Image - Image Transformation
- Scikit Image - Scaling an image
- Scikit Image - Rotating an Image
- Scikit Image - Warping an Image
- Scikit Image - Affine Transform
- Scikit Image - Piecewise Affine Transform
- Scikit Image - ProjectiveTransform
- Scikit Image - EuclideanTransform
- Scikit Image - Radon Transform
- Scikit Image - Line Hough Transform
- Scikit Image - Probabilistic Hough Transform
- Scikit Image - Circular Hough Transforms
- Scikit Image - Elliptical Hough Transforms
- Scikit Image - Polynomial Transform
- Scikit Image - Image Pyramids
- Scikit Image - Pyramid Gaussian Transform
- Scikit Image - Pyramid Laplacian Transform
- Scikit Image - Swirl Transform
- Scikit Image - Morphological Operations
- Scikit Image - Erosion
- Scikit Image - Dilation
- Scikit Image - Black & White Tophat Morphologies
- Scikit Image - Convex Hull
- Scikit Image - Generating footprints
- Scikit Image - Isotopic Dilation & Erosion
- Scikit Image - Isotopic Closing & Opening of an Image
- Scikit Image - Skelitonizing an Image
- Scikit Image - Morphological Thinning
- Scikit Image - Masking an image
- Scikit Image - Area Closing & Opening of an Image
- Scikit Image - Diameter Closing & Opening of an Image
- Scikit Image - Morphological reconstruction of an Image
- Scikit Image - Finding local Maxima
- Scikit Image - Finding local Minima
- Scikit Image - Removing Small Holes from an Image
- Scikit Image - Removing Small Objects from an Image
- Scikit Image - Filters
- Scikit Image - Image Filters
- Scikit Image - Median Filter
- Scikit Image - Mean Filters
- Scikit Image - Morphological gray-level Filters
- Scikit Image - Gabor Filter
- Scikit Image - Gaussian Filter
- Scikit Image - Butterworth Filter
- Scikit Image - Frangi Filter
- Scikit Image - Hessian Filter
- Scikit Image - Meijering Neuriteness Filter
- Scikit Image - Sato Filter
- Scikit Image - Sobel Filter
- Scikit Image - Farid Filter
- Scikit Image - Scharr Filter
- Scikit Image - Unsharp Mask Filter
- Scikit Image - Roberts Cross Operator
- Scikit Image - Lapalace Operator
- Scikit Image - Window Functions With Images
- Scikit Image - Thresholding
- Scikit Image - Applying Threshold
- Scikit Image - Otsu Thresholding
- Scikit Image - Local thresholding
- Scikit Image - Hysteresis Thresholding
- Scikit Image - Li thresholding
- Scikit Image - Multi-Otsu Thresholding
- Scikit Image - Niblack and Sauvola Thresholding
- Scikit Image - Restoring Images
- Scikit Image - Rolling-ball Algorithm
- Scikit Image - Denoising an Image
- Scikit Image - Wavelet Denoising
- Scikit Image - Non-local means denoising for preserving textures
- Scikit Image - Calibrating Denoisers Using J-Invariance
- Scikit Image - Total Variation Denoising
- Scikit Image - Shift-invariant wavelet denoising
- Scikit Image - Image Deconvolution
- Scikit Image - Richardson-Lucy Deconvolution
- Scikit Image - Recover the original from a wrapped phase image
- Scikit Image - Image Inpainting
- Scikit Image - Registering Images
- Scikit Image - Image Registration
- Scikit Image - Masked Normalized Cross-Correlation
- Scikit Image - Registration using optical flow
- Scikit Image - Assemble images with simple image stitching
- Scikit Image - Registration using Polar and Log-Polar
- Scikit Image - Feature Detection
- Scikit Image - Dense DAISY Feature Description
- Scikit Image - Histogram of Oriented Gradients
- Scikit Image - Template Matching
- Scikit Image - CENSURE Feature Detector
- Scikit Image - BRIEF Binary Descriptor
- Scikit Image - SIFT Feature Detector and Descriptor Extractor
- Scikit Image - GLCM Texture Features
- Scikit Image - Shape Index
- Scikit Image - Sliding Window Histogram
- Scikit Image - Finding Contour
- Scikit Image - Texture Classification Using Local Binary Pattern
- Scikit Image - Texture Classification Using Multi-Block Local Binary Pattern
- Scikit Image - Active Contour Model
- Scikit Image - Canny Edge Detection
- Scikit Image - Marching Cubes
- Scikit Image - Foerstner Corner Detection
- Scikit Image - Harris Corner Detection
- Scikit Image - Extracting FAST Corners
- Scikit Image - Shi-Tomasi Corner Detection
- Scikit Image - Haar Like Feature Detection
- Scikit Image - Haar Feature detection of coordinates
- Scikit Image - Hessian matrix
- Scikit Image - ORB feature Detection
- Scikit Image - Additional Concepts
- Scikit Image - Render text onto an image
- Scikit Image - Face detection using a cascade classifier
- Scikit Image - Face classification using Haar-like feature descriptor
- Scikit Image - Visual image comparison
- Scikit Image - Exploring Region Properties With Pandas
Scikit Image - Masked Normalized Cross-Correlation
Masked Normalized Cross-Correlation is an image registration method designed to align images while accommodating the presence of masks or regions of interest within those images. It operates in the Fourier domain using normalized cross-correlation and efficiently incorporates masking to selectively consider or ignore regions of an image during registration. It is well-suited for fast and accurate registration in various computer vision and image analysis applications.
In this tutorial, we use the masked normalized cross-correlation method to determine the relative shift between two similar images containing areas with corrupted or invalid data.
In this context, it's not feasible to apply the masks to the images before computing the cross-correlation, as this would influence the computation. To address this, we need to eliminate the influence of the masks from the cross-correlation results.
Example
In this example, our goal is to register the translation between two images. However, one of these images has approximately 25% of its pixels corrupted.
import numpy as np
import matplotlib.pyplot as plt
from skimage import io
from skimage.registration import phase_cross_correlation
from scipy import ndimage as ndi
# Load an image
image = io.imread('Images/Blue.jpg')
# Define a known shift in pixels
shift = (-22, 13)
# Add the random invalid data
rng = np.random.default_rng()
corrupted_pixels = rng.choice([False, True], size=image.shape, p=[0.25, 0.75])
# Apply the shift to create an offset image with corrupted pixels
offset_image = ndi.shift(image, shift)
offset_image *= corrupted_pixels
print(f'Known offset (row, col): {shift}')
# Create a mask based on the locations of the corrupted pixels
# In this case, the mask is known since we introduced the corruption
mask = corrupted_pixels
# Perform pixel precision image registration
detected_shift = phase_cross_correlation(image, offset_image, reference_mask=mask)
print(f'Detected pixel offset (row, col): {-detected_shift}')
# Create subplots to display results
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, sharex=True, sharey=True, figsize=(8, 3))
ax1.imshow(image, cmap='gray')
ax1.set_axis_off()
ax1.set_title('Reference image')
ax2.imshow(offset_image.real, cmap='gray')
ax2.set_axis_off()
ax2.set_title('Corrupted, offset image')
ax3.imshow(mask, cmap='gray')
ax3.set_axis_off()
ax3.set_title('Masked pixels')
plt.show()
Output
Known offset (row, col): (-22, 13) Detected pixel offset (row, col): [-22. 13.]

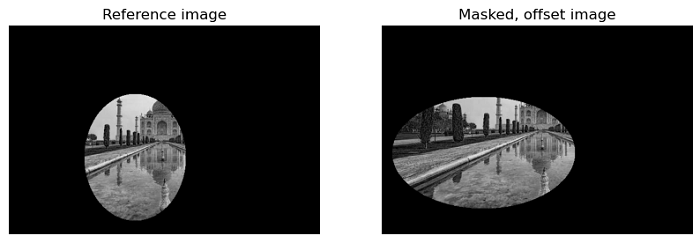
Using Solid masks
Solid masks provide another illustrative example. In this scenario, we have a limited view of both the reference image and an offset image. Notably, the masks applied to these images need not be the same. The phase_cross_correlation function effectively identifies, which part of the images should be compared.
Example
The following example demonstrates the use of the phase_cross_correlation function for pixel-precision image registration while considering solid masks.
import numpy as np
import matplotlib.pyplot as plt
from skimage import io, draw
from skimage.registration import phase_cross_correlation
from scipy import ndimage as ndi
# Load an image
image = io.imread('Images/Blue.jpg')
# Define a known shift in pixels
shift = (-22, 13)
# Create two solid masks using draw.ellipse
rr1, cc1 = draw.ellipse(259, 248, r_radius=125, c_radius=100, shape=image.shape)
rr2, cc2 = draw.ellipse(250, 200, r_radius=110, c_radius=180, shape=image.shape)
# Initialize mask arrays
mask1 = np.zeros_like(image, dtype=bool)
mask2 = np.zeros_like(image, dtype=bool)
# Set the corresponding pixels to True in the masks
mask1[rr1, cc1] = True
mask2[rr2, cc2] = True
# Apply the shift to create an offset image
offset_image = ndi.shift(image, shift)
# Apply masks to the images
image *= mask1
offset_image *= mask2
print(f'Known offset (row, col): {shift}')
# Perform pixel precision image registration with masking
detected_shift = phase_cross_correlation(image, offset_image,
reference_mask=mask1,
moving_mask=mask2)
print(f'Detected pixel offset (row, col): {-detected_shift}')
# Create subplots to display the reference image and the masked, offset image
fig = plt.figure(figsize=(10, 5))
ax1 = plt.subplot(1, 2, 1)
ax2 = plt.subplot(1, 2, 2, sharex=ax1, sharey=ax1)
ax1.imshow(image, cmap='gray')
ax1.set_axis_off()
ax1.set_title('Reference image')
ax2.imshow(offset_image.real, cmap='gray')
ax2.set_axis_off()
ax2.set_title('Masked, offset image')
plt.show()
Output
Known offset (row, col): (-22, 13) Detected pixel offset (row, col): [-22. 13.]