- Scikit Image – Introduction
- Scikit Image - Image Processing
- Scikit Image - Numpy Images
- Scikit Image - Image datatypes
- Scikit Image - Using Plugins
- Scikit Image - Image Handlings
- Scikit Image - Reading Images
- Scikit Image - Writing Images
- Scikit Image - Displaying Images
- Scikit Image - Image Collections
- Scikit Image - Image Stack
- Scikit Image - Multi Image
- Scikit Image - Data Visualization
- Scikit Image - Using Matplotlib
- Scikit Image - Using Ploty
- Scikit Image - Using Mayavi
- Scikit Image - Using Napari
- Scikit Image - Color Manipulation
- Scikit Image - Alpha Channel
- Scikit Image - Conversion b/w Color & Gray Values
- Scikit Image - Conversion b/w RGB & HSV
- Scikit Image - Conversion to CIE-LAB Color Space
- Scikit Image - Conversion from CIE-LAB Color Space
- Scikit Image - Conversion to luv Color Space
- Scikit Image - Conversion from luv Color Space
- Scikit Image - Image Inversion
- Scikit Image - Painting Images with Labels
- Scikit Image - Contrast & Exposure
- Scikit Image - Contrast
- Scikit Image - Contrast enhancement
- Scikit Image - Exposure
- Scikit Image - Histogram Matching
- Scikit Image - Histogram Equalization
- Scikit Image - Local Histogram Equalization
- Scikit Image - Tinting gray-scale images
- Scikit Image - Image Transformation
- Scikit Image - Scaling an image
- Scikit Image - Rotating an Image
- Scikit Image - Warping an Image
- Scikit Image - Affine Transform
- Scikit Image - Piecewise Affine Transform
- Scikit Image - ProjectiveTransform
- Scikit Image - EuclideanTransform
- Scikit Image - Radon Transform
- Scikit Image - Line Hough Transform
- Scikit Image - Probabilistic Hough Transform
- Scikit Image - Circular Hough Transforms
- Scikit Image - Elliptical Hough Transforms
- Scikit Image - Polynomial Transform
- Scikit Image - Image Pyramids
- Scikit Image - Pyramid Gaussian Transform
- Scikit Image - Pyramid Laplacian Transform
- Scikit Image - Swirl Transform
- Scikit Image - Morphological Operations
- Scikit Image - Erosion
- Scikit Image - Dilation
- Scikit Image - Black & White Tophat Morphologies
- Scikit Image - Convex Hull
- Scikit Image - Generating footprints
- Scikit Image - Isotopic Dilation & Erosion
- Scikit Image - Isotopic Closing & Opening of an Image
- Scikit Image - Skelitonizing an Image
- Scikit Image - Morphological Thinning
- Scikit Image - Masking an image
- Scikit Image - Area Closing & Opening of an Image
- Scikit Image - Diameter Closing & Opening of an Image
- Scikit Image - Morphological reconstruction of an Image
- Scikit Image - Finding local Maxima
- Scikit Image - Finding local Minima
- Scikit Image - Removing Small Holes from an Image
- Scikit Image - Removing Small Objects from an Image
- Scikit Image - Filters
- Scikit Image - Image Filters
- Scikit Image - Median Filter
- Scikit Image - Mean Filters
- Scikit Image - Morphological gray-level Filters
- Scikit Image - Gabor Filter
- Scikit Image - Gaussian Filter
- Scikit Image - Butterworth Filter
- Scikit Image - Frangi Filter
- Scikit Image - Hessian Filter
- Scikit Image - Meijering Neuriteness Filter
- Scikit Image - Sato Filter
- Scikit Image - Sobel Filter
- Scikit Image - Farid Filter
- Scikit Image - Scharr Filter
- Scikit Image - Unsharp Mask Filter
- Scikit Image - Roberts Cross Operator
- Scikit Image - Lapalace Operator
- Scikit Image - Window Functions With Images
- Scikit Image - Thresholding
- Scikit Image - Applying Threshold
- Scikit Image - Otsu Thresholding
- Scikit Image - Local thresholding
- Scikit Image - Hysteresis Thresholding
- Scikit Image - Li thresholding
- Scikit Image - Multi-Otsu Thresholding
- Scikit Image - Niblack and Sauvola Thresholding
- Scikit Image - Restoring Images
- Scikit Image - Rolling-ball Algorithm
- Scikit Image - Denoising an Image
- Scikit Image - Wavelet Denoising
- Scikit Image - Non-local means denoising for preserving textures
- Scikit Image - Calibrating Denoisers Using J-Invariance
- Scikit Image - Total Variation Denoising
- Scikit Image - Shift-invariant wavelet denoising
- Scikit Image - Image Deconvolution
- Scikit Image - Richardson-Lucy Deconvolution
- Scikit Image - Recover the original from a wrapped phase image
- Scikit Image - Image Inpainting
- Scikit Image - Registering Images
- Scikit Image - Image Registration
- Scikit Image - Masked Normalized Cross-Correlation
- Scikit Image - Registration using optical flow
- Scikit Image - Assemble images with simple image stitching
- Scikit Image - Registration using Polar and Log-Polar
- Scikit Image - Feature Detection
- Scikit Image - Dense DAISY Feature Description
- Scikit Image - Histogram of Oriented Gradients
- Scikit Image - Template Matching
- Scikit Image - CENSURE Feature Detector
- Scikit Image - BRIEF Binary Descriptor
- Scikit Image - SIFT Feature Detector and Descriptor Extractor
- Scikit Image - GLCM Texture Features
- Scikit Image - Shape Index
- Scikit Image - Sliding Window Histogram
- Scikit Image - Finding Contour
- Scikit Image - Texture Classification Using Local Binary Pattern
- Scikit Image - Texture Classification Using Multi-Block Local Binary Pattern
- Scikit Image - Active Contour Model
- Scikit Image - Canny Edge Detection
- Scikit Image - Marching Cubes
- Scikit Image - Foerstner Corner Detection
- Scikit Image - Harris Corner Detection
- Scikit Image - Extracting FAST Corners
- Scikit Image - Shi-Tomasi Corner Detection
- Scikit Image - Haar Like Feature Detection
- Scikit Image - Haar Feature detection of coordinates
- Scikit Image - Hessian matrix
- Scikit Image - ORB feature Detection
- Scikit Image - Additional Concepts
- Scikit Image - Render text onto an image
- Scikit Image - Face detection using a cascade classifier
- Scikit Image - Face classification using Haar-like feature descriptor
- Scikit Image - Visual image comparison
- Scikit Image - Exploring Region Properties With Pandas
Scikit Image - GLCM Texture Features
Gray-level co-occurrence matrices, often abbreviated as GLCMs, represent a fundamental concept in the field of image processing and computer vision. They serve as a powerful tool for analyzing and characterizing the complex textures present in digital images. Texture is a crucial visual attribute, plays a significant role in various applications, including object recognition, medical imaging, remote sensing, and more.
GLCMs is a histogram of co-occurring grayscale values at a given offset over an image.
The scikit image library has two functions, namely graycomatrix() and graycoprops() for calculating the gray-level co-occurrence matrix and extracting texture properties from a GLCM, respectively.
Using the skimage.feature.graycomatrix() function
The skimage.feature.graycomatrix() function is used for calculating the gray-level co-occurrence matrix (GLCM) from an input image.
Syntax
skimage.feature.graycomatrix(image, distances, angles, levels=None, symmetric=False, normed=False)
Parameters
Here are explanations of the parameters for the function −
image (array_like): It takes an input image, which should be of integer type. And it supports only positive-valued images. If the image type is not uint8, the argument levels needs to be set.
distances (array_like): A list of pixel pair distance offsets.
angles (array_like): A list of pixel pair angles in radians. It specifies the angles at which co-occurrence matrix will be computed.
levels (int, optional): This parameter requires 16-bit images or higher. It indicates the number of gray levels counted, typically 256 for an 8-bit image. The output matrix size is at least levels x levels. In some cases, it's preferable to use binning of the input image rather than large values for levels. The input image should contain integers in [0, levels-1], where levels indicate the number of gray-levels counted (typically 256 for an 8-bit image).
symmetric (bool, optional): If set to True, the output matrix P[:, :, d, theta] is made symmetric. This means it ignores the order of value pairs, so both (i, j) and (j, i) are accumulated when (i, j) is encountered for a given offset. The default value is False.
normed (bool, optional): When True, it normalizes each matrix P[:, :, d, theta] by dividing it by the total number of accumulated co-occurrences for the given offset. This ensures that the elements of the resulting matrix sum to 1. The default value is False.
The function returns a 4-Dimensional ndarray representing the gray-level co-occurrence histogram. The value P[i, j, d, theta] represents the number of times that gray-level j occurs at a distance d and at an angle theta from gray-level i. If normed is False, the output is of type uint32; otherwise, it is float64. The dimensions of the output matrix are levels x levels x number of distances x number of angles.
Example
Let's compute two Gray-Level Co-occurrence Matrices (GLCMs) for an input image. The first GLCM is calculated with a 1-pixel offset to the right, and the second GLCM is computed with a 1-pixel offset upwards.
import numpy as np
from skimage.feature import graycomatrix
# Define the input image
image = np.array([[0, 0, 1, 1],
[0, 0, 1, 1],
[0, 2, 2, 2],
[2, 2, 3, 3]], dtype=np.uint8)
# Calculate the GLCM with specified parameters
result = graycomatrix(image, [1], [0, np.pi/4, np.pi/2, 3*np.pi/4], levels=4)
# Display the resulting GLCMs
print('Theta = 0')
print(result[:, :, 0, 0])
print('Theta = 1')
print(result[:, :, 0, 1])
print('Theta = 2')
print(result[:, :, 0, 2])
print('Theta = 3')
print(result[:, :, 0, 3])
Output
Theta = 0 [[2 2 1 0] [0 2 0 0] [0 0 3 1] [0 0 0 1]] Theta = 1 [[1 1 3 0] [0 1 1 0] [0 0 0 2] [0 0 0 0]] Theta = 2 [[3 0 2 0] [0 2 2 0] [0 0 1 2] [0 0 0 0]] Theta = 3 [[2 0 0 0] [1 1 2 0] [0 0 2 1] [0 0 0 0]]
Using the skimage.feature.graycoprops() function
The skimage.feature.graycoprops() function is used to calculate various texture properties of a Gray-Level Co-occurrence Matrix (GLCM). Calculate a characteristic of a gray-level co-occurrence matrix to provide a compact summary of the matrix's properties. The properties are computed as follows −
contrast: $\mathrm{\sum_{i,j=0}^{levels-1} P_{i,j} (i-j)^2}$
dissimilarity: $\mathrm{\sum_{i,j=0}^{levels-1} P_{i,j} \lvert i-j\rvert}$
homogeneity: $\mathrm{\sum_{i,j=0}^{levels-1} \frac{P_{i,j}}{1+(i-j)^2}}$
ASM: $\mathrm{\sum_{i,j=0}^{levels-1} P_{i,j}^{2}}$
energy: $\mathrm{\sqrt{ASM}}$
correlation:$\mathrm{\sum_{i,j=0}^{levels-1}P_{i,j}\left[\frac{(i-\mu_{i})(j-\mu_{i})}{\sqrt{(\sigma_{i2})(\sigma_{j2})}} \right ]}$
Syntax
Here is the syntax of this method −
skimage.feature.graycoprops(P, prop='contrast')
Parameters
The method accepts the following parameters −
P (ndarray): This parameter takes the input array, which is the gray-level co-occurrence histogram (GLCM) for which to compute the specified property. The value P[i, j, d, theta] represents the number of times that gray-level j occurs at a distance d and at an angle theta from gray-level i.
prop (str, optional): It specifies the property of the GLCM to compute. It accepts the following options: 'contrast', 'dissimilarity', 'homogeneity', 'ASM', 'energy', 'correlation'. The default value is 'contrast'.
The function returns a 2-dimensional array where results[d, a] represents the property specified by prop for the d-th distance and the a-th angle.
Each GLCM is normalized to have a sum of 1 before the computation of these texture properties.
Example
The following example calculates the contrast properties from the gray-level co-occurrence matrix (GLCM) using the graycoprops() function.
import numpy as np
from skimage.feature import graycomatrix, graycoprops
# Define the input image
image = np.array([[0, 0, 1, 1],
[0, 0, 1, 1],
[0, 2, 2, 2],
[2, 2, 3, 3]], dtype=np.uint8)
# Calculate the gray-level co-occurrence matrix (GLCM)
g = graycomatrix(image, [1, 2], [0, np.pi/2], levels=4,
normed=True, symmetric=True)
# Compute the 'contrast' property from the GLCM
contrast = graycoprops(g, 'contrast')
# Print the 'contrast' property
print("Contrast:", contrast[0, 0])
Output
Contrast: 0.5833333333333333
Example
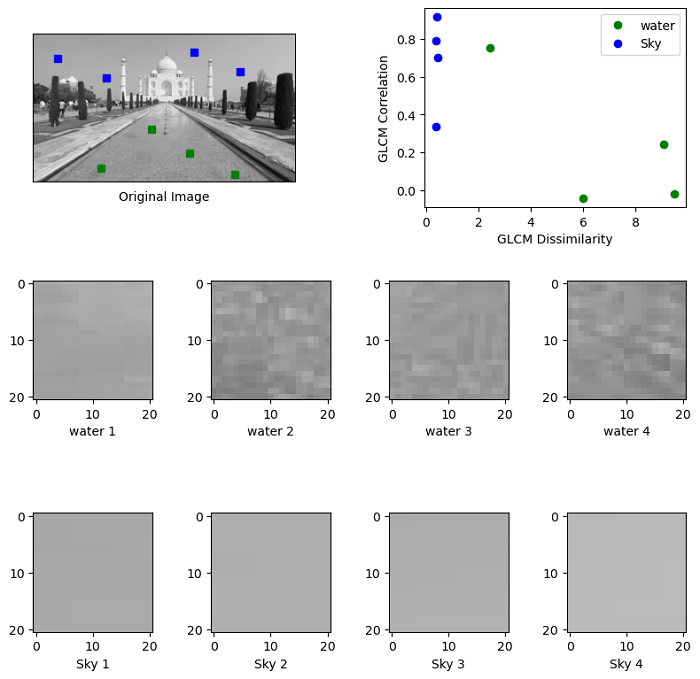
This example demonstrates texture classification using the utilization of Gray Level Co-occurrence Matrices (GLCMs). Specifically, the example focuses on distinguishing between two distinct textures present in an image: water areas and sky areas.
For each selected patch within the image, a GLCM is computed with a horizontal offset of 5 (specified as distance=[5] and angles=[0]). Subsequently, two critical features, namely dissimilarity and correlation, are extracted from these GLCM matrices.
import matplotlib.pyplot as plt
from skimage.feature import graycomatrix, graycoprops
from skimage import io, util, exposure
PATCH_SIZE = 21
# open the input image
image = util.img_as_ubyte(io.imread('Images/Tajmahal_3.jpg', as_gray=True))
image = exposure.rescale_intensity(image)
# select some patches from water areas of the image
water_locations = [(160, 200), (230, 110), (202, 268), (240, 350)]
water_patches = []
for loc in water_locations:
water_patches.append(image[loc[0]:loc[0] + PATCH_SIZE,
loc[1]:loc[1] + PATCH_SIZE])
# select some patches from sky areas of the image
sky_locations = [(33, 33), (68, 119), (22, 276), (56, 359)]
sky_patches = []
for loc in sky_locations:
sky_patches.append(image[loc[0]:loc[0] + PATCH_SIZE,
loc[1]:loc[1] + PATCH_SIZE])
# compute some GLCM properties each patch
xs = []
ys = []
for patch in (water_patches + sky_patches):
glcm = graycomatrix(patch, distances=[5], angles=[0], levels=256,
symmetric=True, normed=True)
xs.append(graycoprops(glcm, 'dissimilarity')[0, 0])
ys.append(graycoprops(glcm, 'correlation')[0, 0])
# create the figure
fig = plt.figure(figsize=(8, 8))
# display original image with locations of patches
ax = fig.add_subplot(3, 2, 1)
ax.imshow(image, cmap=plt.cm.gray,
vmin=0, vmax=255)
for (y, x) in water_locations:
ax.plot(x + PATCH_SIZE / 2, y + PATCH_SIZE / 2, 'gs')
for (y, x) in sky_locations:
ax.plot(x + PATCH_SIZE / 2, y + PATCH_SIZE / 2, 'bs')
ax.set_xlabel('Original Image')
ax.set_xticks([])
ax.set_yticks([])
ax.axis('image')
# for each patch, plot (dissimilarity, correlation)
ax = fig.add_subplot(3, 2, 2)
ax.plot(xs[:len(water_patches)], ys[:len(water_patches)], 'go',
label='water')
ax.plot(xs[len(water_patches):], ys[len(water_patches):], 'bo',
label='Sky')
ax.set_xlabel('GLCM Dissimilarity')
ax.set_ylabel('GLCM Correlation')
ax.legend()
# display the image patches
for i, patch in enumerate(water_patches):
ax = fig.add_subplot(3, len(water_patches), len(water_patches)*1 + i + 1)
ax.imshow(patch, cmap=plt.cm.gray,
vmin=0, vmax=255)
ax.set_xlabel(f"water {i + 1}")
for i, patch in enumerate(sky_patches):
ax = fig.add_subplot(3, len(sky_patches), len(sky_patches)*2 + i + 1)
ax.imshow(patch, cmap=plt.cm.gray,
vmin=0, vmax=255)
ax.set_xlabel(f"Sky {i + 1}")
plt.tight_layout()
plt.show()
Output